| enginuity |
|
3D Ultrasound
It may soon be possible to use ultrasound more widely for medical diagnoses, including some cases where MRI and CT scanners were required, owing to research being carried out by a team led by Dr Richard Prager and Dr Andrew Gee. Conventional ultrasound machines, currently in routine use in many clinics, are limited to producing two-dimensional slices through the area being examined.
Clinicians are often more interested in making volume measurements, however. The team's work is directed towards making this possible by extending the capabilities of conventional ultrasound machines. 'We are upgrading a cheap accessible technique, ultrasound, to replace the expensive and invasive techniques of CT and MRI scanning,' explains Dr Prager.
Essentially, their technique involves using a computer to build up a 3D image from a series of 2D slices. To do this, precise information is needed on the position and orientation of the ultrasound probe as it is moved over the area of interest. Position sensor technology, coming out of the virtual reality industry, has been harnessed and combined with a unique calibration method to solve this problem.
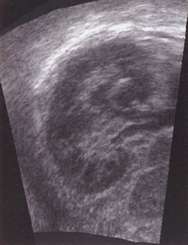
This coronal view of a human kidney was obtained by reslicing a 3D ultrasound data set acquired in a different direction. The coronal view is impossible to obtain using standard 2D ultrasound, since the ribs get in the way.
Dr Prager explains: 'It has been difficult to calibrate 3D ultrasound systems with sufficient accuracy, since we are trying to measure the relative position and orientation of the position sensor, which we attach to the probe's casing, and the ultrasound beam, which is invisible.' The team has solved this problem using a relatively simple but ingenious device, which has been patented through Cambridge University Technical Services (CUTS).
No 'fudge factors'
As well as solving the calibration problem, the team has also developed new ways of 'reslicing' the data set. This produces a 2D image which would have been impossible to obtain using conventional ultrasound (because the slice lies entirely below the skin, for instance). 'We work directly from raw data, and can produce the reslice image immediately after, or even during, the ultrasound examination,' explains Dr Gee. 'Unlike existing techniques, our approach requires no 'fudge factors' or further processing of the data, so we can obtain high-quality images without any delay.'
Graham Treece, a first-year research student working in the team, has developed a new algorithm capable of inferring 3D surfaces from multi-axial slices. This allows anatomical shapes to be reconstructed from contours, which are traced out by the clinician in the individual 2D slices and are not necessarily parallel. The surface can then be visualised and its volume calculated much more accurately than is possible using alternative techniques.
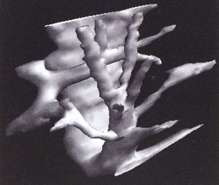
Following segmentation of the 3D ultrasound data set, it is possible to visualise anatomical surfaces and calculate the enclosed volumes. This example shows the system of hepatic veins inside a human liver.
The engineering team works in close collaboration with Dr Laurence Berman, a consultant radiologist at Addenbrooke's Hospital.
Further details are available on the team's website: http://svr-www.eng.cam.ac.uk/~rwp/stradx/ or from Dr Richard Prager, tel: 01223 332771 or Dr Andrew Gee, tel: 01223 332750.
| number 7, December '98 |